By: Sherman McRae
Discover how African mtDNA haplogroups reveal your maternal ancestry, from humanity’s oldest lineages to powerful connections across the African diaspora.
This Juneteenth, as we celebrate freedom and honor the memory of our ancestors, there’s perhaps no more profound way to reconnect with your heritage than through the genetic legacy carried within your very cells. Your mitochondrial DNA (mtDNA) preserves an unbroken chain of ancestry that links you directly to your African foremothers—telling an intimate story of resilience, migration, and survival that spans tens of thousands of years and transcends the trauma of the Middle Passage.
How Mitochondrial DNA Connects You to Your African Ancestry
Mitochondrial DNA is unique because it’s passed down exclusively through the maternal line—from mother to child, generation after generation, largely unchanged. This means the mtDNA you carry today is virtually identical to that of your direct maternal ancestor who lived in Africa hundreds or even thousands of years ago.
Haplogroup L and the Major African Maternal Lineages That Shaped Human History
Before diving into specific haplogroups, it’s important to understand how maternal lineages are categorized. These lineages are grouped into what scientists call haplogroups. These are genetic branches that trace back to a single common ancestor thousands of years ago. In Africa, these haplogroups begin with the letter L, representing the most ancient maternal lines in human history.
Each African haplogroup represents a unique thread in the broader story of human evolution, migration, and adaptation. From the earliest hunter-gatherers in southern Africa to the great Bantu migrations and connections across the Indian Ocean, these genetic markers allow us to follow the paths our foremothers walked, sometimes thousands of years before recorded history.
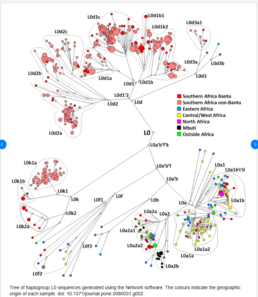
Haplogroup L0: Humanity’s Oldest Maternal Lineages
Mitochondrial Eve’s oldest daughter L0 was born around 121,000 years ago (121kya). mtDNA Haplogroup L0 represents some of humanity’s most ancient maternal lineages.
This haplogroup reaches its highest frequencies among the Khoisan peoples of southern Africa, with some populations showing remarkable concentrations among:
The L0 haplogroup consists of five main branches—L0a, L0b, L0d, L0f, and L0k—each with distinct geographic distributions that tell unique stories of ancient human migrations and population histories.
L0a: A Widely Spread Lineage with Surprising Geographic Reach
L0a displays the broadest geographic distribution of all L0 subclades, appearing throughout southeastern Africa with notable presence in Mozambique. Remarkably, L0a also appears in unexpected locations: it has a significant presence in Hadramawt, Yemen, and is found among Guineans in West Africa, with the Balanta group showing elevated frequencies. L0a is also commonly observed among the Mbuti and Biaka Pygmies of Central Africa.
L0b: East Africa’s Branch of the Earliest African Lineage
L0b is primarily found in Ethiopia, representing the Horn of Africa’s contribution to this ancient maternal lineage. If you carry L0, your maternal line traces back to some of the earliest human populations, connecting you to the deepest roots of human ancestry in Africa.
L0d, L0f, and L0k: Southern Africa’s Ancient and Isolated Subclades
L0d and L0k subclades are almost exclusively restricted to the Khoisan peoples of southern Africa, though L0d has also been detected among the Sandawe people of Tanzania, suggesting an ancient connection between southern African and East African click-speaking populations.
L0f is found in relatively small frequencies among the Sandawe people of Tanzania in East Africa, representing another ancient East African lineage.
Haplogroup L1: An Ancient Lineage Rooted in Central and West Africa
Haplogroup L1 represents ancient maternal lineages that emerged over 100,000 years ago (100 kya). Most common in Central Africa and West Africa, L1 is primarily found among forest populations and was likely once more widespread before being constrained to its current distribution by later population movements, particularly the Bantu expansion.
The haplogroup consists of two main branches—L1b and L1c—each with distinct geographic patterns and population associations.
L1c: A Forest-Dwelling Lineage from the Atlantic to Central Africa
L1c, dating to approximately 72,000 years ago (72 kya) reaches its highest frequency in West and Central Africa. This subclade shows remarkable concentrations among Central African populations including the Ba-Kola, and Biaka peoples, as well as the Tikar, Baka peoples of Gabon and Cameroon, the Bakoya, and Ba-Bongo groups.
L1c is also commonly found in São Tomé and Angola, and occurs frequently among Central African Bantu speakers, having likely originated among peoples near the Atlantic coast in western equatorial Africa.
L1b: A Younger West African Lineage That Traveled North
L1b, a much more recent lineage dating to about 32,000 years ago (32kya), is concentrated in western Africa but also extends into central and northern Africa. While showing its highest diversity in West Africa’s Bight of Biafra region, L1b has spread northward through trans-Saharan migration routes into North Africa.
If you carry L1, your maternal line traces back to the ancient forest populations of Central and West Africa, connecting you to some of the continent’s earliest settled communities and their rich cultural traditions.
Haplogroup L2: A Central Player in African Migration and Maternal Legacy
Haplogroup L2 originated in Western Africa approximately 80,000 years ago (80 kya) and now has the most widespread distribution of any African maternal lineage, found throughout sub-Saharan Africa and strongly associated with the Bantu expansion.
This haplogroup represents one of the most successful maternal lineages in African history, spreading from its West African origins to cover virtually the entire continent south of the Sahara.
L2 consists of five main subclades, each with distinct geographic patterns that trace different waves of migration and population movement across Africa.
- L2a
- L2b
- L2c
- L2d
- L2e
L2a: Sub-Saharan Africa’s Most Common mtDNA Lineage
L2a, dating to approximately 71,000 years ago (71 kya), is the most common and widely distributed sub-Saharan African haplogroup, found from West Africa to East Africa and throughout the continent.
This subclade shows particularly high frequencies in:
- Chad
- Kenya
- Uganda
- Tanzania
- Mozambique
- Ghana
L2a extends into North Africa among populations in:
- Egypt
- Morocco
- Algeria
- Tunisia
L2a also reaches into the Arabian Peninsula, appearing in Yemen and other Middle Eastern populations.
The L2a2 subclade is specifically characteristic of the Mbuti Pygmies of Central Africa, while various L2a subclades show clear evidence of both ancient migrations into East Africa and more recent Bantu expansion movements.
L2b, L2c, and L2d: West African Lineages That Reached Europe and Beyond
L2b, L2c, and L2d are primarily concentrated in West Africa, where they likely originated, though they have spread to varying degrees across the continent.
L2b appears throughout West and Central Africa and has even reached Europe, with specific subclades found in Spain and Italy.
L2c shows its highest frequencies in Senegal, Cape Verde, and Guinea-Bissau, with some presence extending to North Africa and Europe.
L2d remains largely restricted to West Africa but also appears in Yemen, Mozambique, and Sudan, indicating ancient trade connections.
L2e: A West African Lineage with Deep Regional Roots
L2e is typical of West Africa, particularly among the Mandinka people of Guinea-Bissau, and is also found in Tunisia and among African American populations.
The L2 haplogroup, with most Southern and Eastern African L2 lineages showing recent origins from Central African clades within the last few millennia.
These lineages display characteristic “star-like” patterns indicating rapid population expansion and migration. If you carry L2, your maternal line connects you to one of Africa’s great population movements, linking you to the agricultural communities that transformed the continent and carried their cultural and genetic legacy from the Atlantic coast to the Indian Ocean.
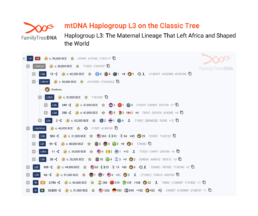
Haplogroup L3: The Maternal Lineage That Left Africa and Shaped the World
Haplogroup L3 holds a unique place in human history as the ancestral lineage of all non-African populations, yet it also represents one of the most diverse and widespread maternal lineages within Africa itself.
Originating in East Africa approximately 60,000 years ago (60 kya), L3 played a pivotal role in the out-of-Africa migration that populated the entire world, while simultaneously expanding across the African continent through multiple waves of migration. This haplogroup encompasses numerous subclades, each telling distinct stories of ancient movements, trade routes, and population expansions that have shaped African genetic diversity for tens of thousands of years.
L3’s East African origins are evident in several subclades that remain virtually exclusive to this region.
L3a and L3h are found almost entirely in Ethiopia, Kenya, Tanzania, and Uganda, representing the most ancient East African maternal lineages.
L3i and L3x are similarly restricted to East Africa, with L3i extending into the Horn of Africa and reaching Yemen and Oman, while L3x appears among Ethiopian Oromos and extends northward into Egypt. These eastern lineages preserve the genetic signatures of the populations from which all modern humans outside Africa ultimately descended.
The most common L3 lineages in Central and West Africa—L3b, L3d, and L3e—tell powerful stories of population movements and cultural transformations.
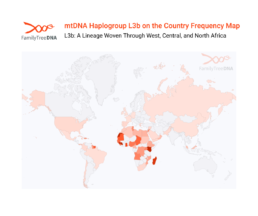
L3b: A Lineage Woven Through West, Central, and North Africa
L3b dominates across West and Central Africa, from Nigeria and Ghana to Cameroon and the Democratic Republic of Congo, and has spread through trans-Saharan trade routes into North Africa.
L3d: A Sahel-to-Nile Lineage with Reach from Ghana to Yemen
L3d is widespread among the Fulani pastoralists, Chadian populations, and Akan peoples of Ghana, extending from the Sahel to Mozambique and even reaching Yemen and Egypt.
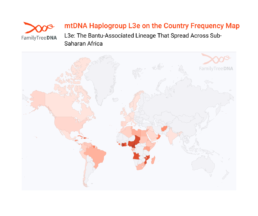
L3e: The Bantu-Associated Lineage That Spread Across Sub-Saharan Africa
L3e, the most common L3 subclade among Bantu-speaking populations, spreading from its Central African origins to become the predominant maternal lineage across much of sub-Saharan Africa.
Haplogroups L4, L5, and L6: Rare East African Lineages from Humanity’s Earliest Roots
Haplogroups L4, L5, and L6 represent ancient East African maternal lineages that provide crucial insights into early human populations in the region. These haplogroups, dating between 22,000-120,000 years ago (22-121kya), remained primarily within East Africa rather than participating in global migrations like L3.
These lesser-known haplogroups didn’t contribute to major global migrations but remain vital to understanding the early diversification of maternal lineages in Africa.
L4 and L6 are particularly concentrated in the Horn of Africa and eastern Rift Valley, while L5 represents one of the most ancient lineages, found among populations considered to be among the earliest divergent human groups.
L4, arising around 70,000 years ago (70kya), shows its highest frequencies in Tanzania, particularly among the Sandawe and Hadza populations, representing some of the most ancient hunter-gatherer traditions in East Africa.
L6, dating to approximately 22,000 years ago (22 kya), is primarily limited to the Horn of Africa and southern East Africa, with some subclades extending into Saudi Arabia.
L5: An Ancient Lineage Linking East Africa to the Arabian Peninsula
L5, emerging around 100,000 years ago (100 kya), reaches its peak frequencies among the Mbuti Pygmies of the eastern Democratic Republic of Congo, the Tshwa and Shua Khoisan of Botswana, and the Sandawe of Tanzania. This ancient lineage has even been detected in Saudi Arabia, indicating early connections between East Africa and the Arabian Peninsula.
Haplogroup L7: A Newly Discovered African Lineage That Rewrites Maternal History
Recent discoveries continue to reshape our understanding of African maternal lineages. In 2022, researchers working with FamilyTreeDNA’s Million Mito Team identified L7, a previously unknown 100,000-year-old maternal lineage that had been misclassified for years. This groundbreaking discovery, made possible by analyzing over 200,000 complete mtDNA sequences in FamilyTreeeDNA’s database, added an entirely new branch to the human family tree and highlighted how much we still have to learn about African genetic diversity.
The L7 lineage, found primarily among the Sandawe people of Tanzania, represents one of the oldest maternal lineages ever discovered, with its most ancient subclade L7a* dating back approximately 80,000 years (80 kya).
This discovery demonstrates that African populations continue to hold keys to understanding our earliest human heritage and shows how advanced genetic testing and large-scale databases are revolutionizing our knowledge of human origins.
mtDNA Haplogroups That Came Back: Tracing Ancient Migrations into Africa
While the vast majority of African and African diaspora populations carry L haplogroups that originated on the continent, some individuals may discover they belong to non-African maternal lineages that entered Africa through ancient migrations and historical movements. These haplogroups tell fascinating stories of population exchanges, trade routes, and cultural interactions that have shaped African genetic diversity over tens of thousands of years.
The most significant of these non-African lineages are haplogroups U6, B4a1a1b, and several others that represent ancient “back-to-Africa” migrations from Asia and Europe.
These non-African haplogroups are particularly common in North Africa, East Africa, and Madagascar, but can also be found among populations throughout sub-Saharan Africa due to ancient migrations and more recent gene flow. For African Americans and others in the diaspora, discovering one of these lineages can open unexpected windows into complex migration histories that connect Africa to the broader human story.
Understanding these lineages requires looking beyond Africa’s borders to trace the journeys that brought these maternal lines to the continent, where they became integrated into African communities and cultures.
Haplogroup U6a5: A Back-to-Africa Lineage Rooted in North and West Africa
U6a5 represents a significant subclade of the North African haplogroup U6 that demonstrates a clear pattern of West African spread. The broader U6 haplogroup represents an ancient “back-to-Africa” migration from southwestern Asia that established itself in North Africa around 30,000 years ago (30 kya).
U6a5 specifically traces the southward migration routes from this North African base through the Sahel into West Africa, making it distinct from other U6 subclades that remained in the Mediterranean region or spread into Europe.
Timeline and Migration: How U6a5 Became Indigenous to West Africa
The U6a5 lineage dates back approximately 10,000 years ago (10kya) but its most significant expansion occurred much later when autochthonous (indigenous, from the Greek word autokhthon, meaning literally “sprung from the land itself”) clusters in sub-Saharan Africa first appeared within U6a5b.
Researchers suggest this migration could have been due to climactic changes over time forcing populations to seek more hospitable regions to live.
Key Subclades
- U6a5a: Represents migration pathways from the Sahel into West Africa, with further spreads of secondary U6a branches going southwards to Sahel countries and reaching West Africa.
- U6a5b: Defined by mutations (3714, 16184, 16234), this subclade marked the establishment of truly indigenous West African U6 populations.
U6a5 in the African Diaspora: A Genetic Bridge from North Africa to the Americas
U6a5 has direct relevance to African American and diaspora heritage, as research shows that U.S. sequences are found within a mainly sub-Saharan Africa background (U6a3c, U6a5). The broader U6 haplogroup appears among African Americans and people of the diaspora, providing a direct maternal link between modern populations and ancient West African communities that descended from the North African U6 expansion thousands of years ago.
This lineage exemplifies how ancient population movements from North Africa contributed to West African genetic diversity and, through the transatlantic slave trade, became part of African American and diaspora genetic heritage.
mtDNA Haplogroups from Madagascar: Southeast Asian Roots in the African Diaspora
Madagascar presents one of the most fascinating examples of genetic admixture in human history, where approximately equal African and Indonesian contributions to both paternal and maternal Malagasy lineages have created a unique population with profound implications for understanding African diaspora genetics.
The Malagasy Motif: A Unique mtDNA Signature of African–Asian Fusion
B4a1a1b, or “The “Malagasy Motif”found in the Malagasy people represent a remarkable genetic fusion that occurred relatively recently in human history. Coastal Malagasy populations, including the Temoro, Vezo, and Mikea, have approximately 70% African ancestry and 30% Asian ancestry, while highlander tribes tend to have lower African ancestry at around 45%.
This genetic composition reflects the complex settlement history of Madagascar by both Austronesian-speaking peoples from Southeast Asia and Bantu-speaking populations from East Africa.
This is perhaps the most significant Austronesian maternal lineage found in Madagascar and represents a unique variant of the famous “Polynesian motif.”
The Malagasy Motif is defined by two specific mutations, C1473T and T3423A, that are found in all carriers of this lineage in Madagascar. These two additional mutations distinguish the Malagasy variant from the broader Polynesian motif, creating a Madagascar-specific signature.
Other Malagasy Lineages: Madagascar also hosts several other maternal lineages including but not limited to:
- M7c1c
- M23
- F3b
- B4a1a1
- B4a
These varying lineages reflect the diverse origins of the founding populations.
Malagasy DNA in the Diaspora: A Story of Trade, Migration, and Survival
The relevance of Malagasy haplogroups to African American and the broader African diaspora populations is both historical and genetic. Seventeen known slave ships came from Madagascar to North America during the Transatlantic Slave Trade. As a result, we find Malagasy DNA in the African American descendants of enslaved people, often of Southeast Asian origin.
The presence of these haplogroups among African diaspora populations tells a story that extends far beyond the typical narrative of African ancestry. It connects carriers to the remarkable Austronesian maritime expansion that reached across the Indian Ocean, the complex demographic history of Madagascar, and the often-overlooked diversity of populations affected by the transatlantic slave trade.
Tracing Your Maternal Line Starts Here
At FamilyTreeDNA, every mitochondrial DNA test is a Full Sequence test. That means when you test with us, you’re not just getting a glimpse—you’re getting the complete picture. We analyze all ~16,569 base pairs of your mitochondrial genome, delivering the highest possible resolution for exploring your direct maternal line.
This is the gold standard for maternal ancestry research, trusted by both academic researchers and African-descended individuals around the world who are reclaiming their genetic past.
What You Get with and mtDNA Test
- Complete haplogroup assignment: Receive a precise subclade like L2a1a2a1, not just a broad regional label.
- Full list of maternal mutations: Every defining variant is identified and tracked, giving you deeper insight into your lineage.
- Maternal matching at multiple levels: Connect with, exact matches (identical mtDNA), near matches (1–3 mutations apart), and haplogroup matches (same ancestral branch)
- Lineage timeline and SNP details: See when your maternal line branched and how it fits into human history.
- Automatic research updates: As new subclades are discovered, your results evolve with no retesting required.
Built on the Largest African mtDNA Database in the World
Your results are compared to a global reference that includes:
- Thousands of samples from archaeological sites, university studies, and historic burial grounds
- Modern testers across the African continent, thanks to our ongoing African Research Initiative
- Rare African haplogroups that are often missing from other databases
Our database has helped testers in the diaspora connect with long-lost roots, like a Jamaican man whose mtDNA matched a burial at Maryland’s historic Catoctin Furnace, or a Congolese tester matched to others descended from the Bakongo tribe.
Make the Most of Your Results
- Upload a family tree to connect with maternal matches
- Explore the histories and geographies of your haplogroup
- Join Group Projects that focus on African ancestry
- Revisit your results regularly to see matches grow as the database grows
Your Maternal DNA is a Legacy
The mtFull Sequence test provides the gold standard for maternal ancestry research, especially valuable for those seeking to understand their African heritage with maximum precision. With FamilyTreeDNA’s unparalleled database of academic samples and ongoing research initiatives, your results will only become more detailed and meaningful over time.
Your mtDNA is a genetic time capsule, preserving the essence of your African maternal ancestors within your very cells. This Juneteenth, as we celebrate freedom and honor our heritage, there’s no more powerful way to connect with your roots than through the unbroken maternal line that links you directly to the African continent.
Ready to discover your African maternal heritage? Learn more about our comprehensive mtDNA testing and start your journey of ancestral discovery today.
References
Behar, D. M., et al. (2008). The dawn of human matrilineal diversity. American Journal of Human Genetics, 82, 1130–1140.
Behar, D. M., Villems, R., Soodyall, H., Blue-Smith, J., Pereira, L., Metspalu, E., Scozzari, R., Makkan, H., Tzur, S., Comas, D., Bertranpetit, J., Quintana-Murci, L., Tyler-Smith, C., Wells, R. S., & Rosset, S. (2007). The dawn of human matrilineal diversity. Molecular Biology and Evolution, 24(3), 757-768.
https://doi.org/10.1093/molbev/msl209
FamilyTreeDNA. (2025). Malagasy Roots – Background. FamilyTreeDNA Groups. Retrieved June 10, 2025, from https://www.familytreedna.com/groups/malagasy-dna/about/background
FamilyTreeDNA. (2025). MitoTree: Mitochondrial DNA Phylogenetic Tree. FamilyTreeDNA. https://www.familytreedna.com
Kivisild, T., et al. (2004). Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. American Journal of Human Genetics, 75, 752–770.
Maca-Meyer, N., González, A.M., Pestano, J., et al. (2003). Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genetics, 4, 15.
Maier, R., et al. (2022). [Complete title needed]. Scientific Reports.
Pala, M., et al. (2022). African mitochondrial haplogroup L7: a 100,000-year-old maternal human lineage discovered through reassessment and new sequencing. Scientific Reports, 12, 9455.
Pierron, D., Razafindrazaka, H., Pagani, L., et al. (2017). Genomic landscape of human diversity across Madagascar. Proceedings of the National Academy of Sciences, 114(32), E6498-E6506.
Rosa, A., et al. (2004). MtDNA profile of West Africa Guineans: towards a better understanding of the Senegambia region. Annals of Human Genetics, 68, 340–352.
Salas, A., et al. (2002). The making of the African mtDNA landscape. American Journal of Human Genetics, 71, 1082–1111.
Secher, B., Fregel, R., Larruga, J.M., et al. (2014). The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evolutionary Biology, 14, 109.
Silva, M., et al. (2015). 60,000 years of interactions between Central and Eastern Africa documented by major African mitochondrial haplogroup L2. Scientific Reports, 5, 12526.
Soares, P., et al. (2012). The Expansion of mtDNA Haplogroup L3 within and out of Africa. Molecular Biology and Evolution, 29(3), 915–927.
Torroni, A., et al. (2006). Harvesting the fruit of the human mtDNA tree. Trends in Genetics, 22, 339–345.
Watson, E., Forster, P., Richards, M. & Bandelt, H. J. (1997). Mitochondrial footprints of human expansions in Africa. American Journal of Human Genetics, 61, 691–704.
Additional Sources Consulted
“The First Modern Human Dispersals across Africa” (2013)
“MtDNA control region variation affirms diversity and deep sub-structure in populations from southern Africa” (2013)

About the Author
Sherman McRae
Independent Contractor for FamilyTreeDNA
Sherman is an experienced genealogist specializing in African American genetic genealogy and DNA research. Currently, he serves as a contributor in the FamilyTreeDNA Research & Development Department, where he actively supports DNA research initiatives. Sherman has been invited to deliver presentations at national and local events, sharing his expertise on utilizing DNA testing in genealogical research and participating on panels to discuss strategies for increasing African participation in DNA testing.
Sherman’s passion for genetic genealogy drives his ongoing research and commitment to advancing genetic heritage understanding for African Americans and people of the diaspora. Through his work, he continues to explore innovative methodologies and contribute to the evolving field of DNA-based genealogical research, helping to bridge the gap between cutting-edge genetic science and accessible ancestry discovery for African American communities.